Bru-seq
Overview
Bru-seq refers to a family of related technologies for exploring the dynamics of transcription biology in mammalian cells. The methods are related by the selection and sequencing of only RNA molecules that were metabolically labelled during an exposure of the cells to bromouridine (Bru).
Dr. Wilson has worked for more than a decade with Bru-seq inventor and project leader, Dr. Mats Ljungman, to help develop the associated bioinformatics tools for data analysis. The pipeline and browser developed by our team have served the needs of researchers worldwide.
Most recently we are finalizing publications related to work done within the ENCODE Project , in which a large number of cell lines were subjected to Bru-seq in parallel with other RNA-based methods. The combinatorial data sets have provided novel insights into transcription regulatory control (see figure).
Profiling transciption activity genome wide. The panels hint at the power of parallel assessments of nascent transcription - we now have thousands of such data sets to mine.
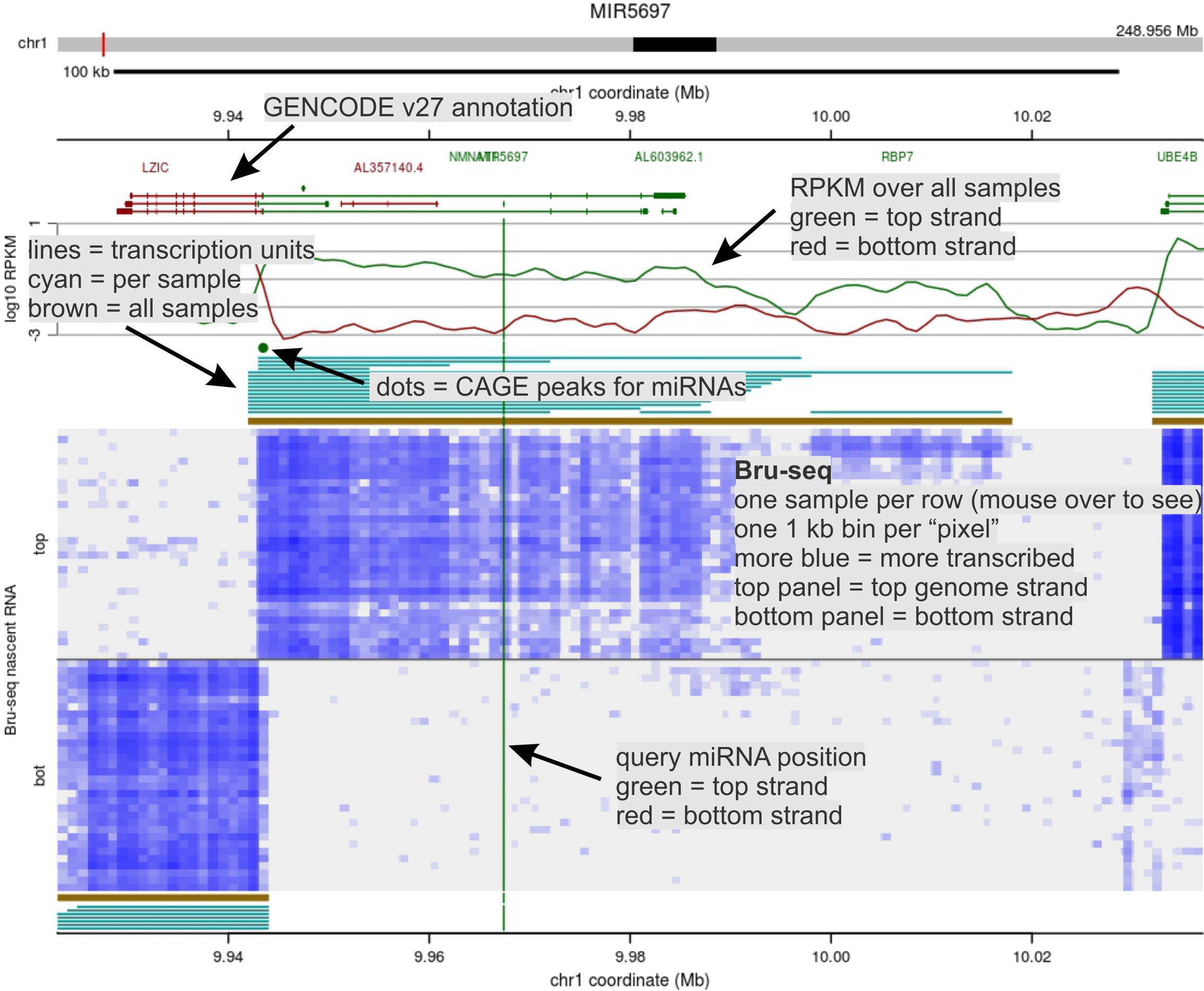
Profiling transciption activity genome wide. The panels hint at the power of parallel assessments of nascent transcription - we now have thousands of such data sets to mine.
Learn more about Bru-Seq
https://rna.umich.edu/research/cores/bru-seq-lab/