Mammalian CNV Formation
Overview
Dr. Wilson has worked for more than a decade with Dr. Tom Glover, an expert cytogeneticist and leader in the study of chromosomal common fragile sites (CFSs). CFSs are regions of chromosomes that are highly susceptible to forming lesions, known as “breaks” and “gaps”, on metaphase chromosomes. They are often used as a hallmark of genomic instability upon replication stress.
Common fragile sites (CFSs). Typical breaks and gaps seen on metaphase chromosomes at CFS loci FRA3B (the FHIT gene) and FRA16D (the WWOX gene) upon replication stress.
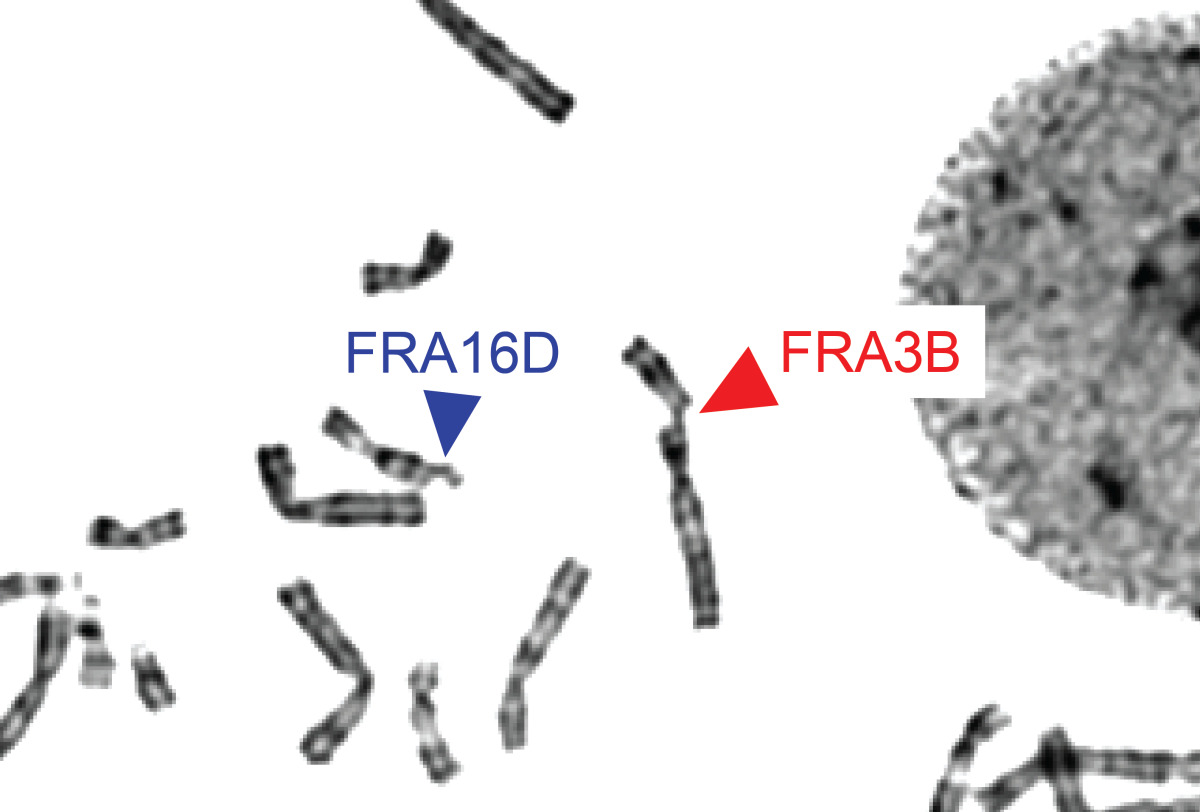
Common fragile sites (CFSs). Typical breaks and gaps seen on metaphase chromosomes at CFS loci FRA3B (the FHIT gene) and FRA16D (the WWOX gene) upon replication stress.
Together we have explored whether CFSs are also subject to mutation accumulation in response to replication stresses, in particular chromosomal rearrangements in the form of copy number variants (CNVs) or other structural variants (SVs). Indeed they are, but with a specific pattern where hotspots of mainly deletion CNV formation are observed in a subset of human genes that are extremely large (>0.5 Mb). Over a series of collaborative papers we have shown that many forms of replication stress will induce this CNV pattern, but only at large genes that are actively transcribed.
We also presented data consistent with other literature that transcription causes CFS genes to undergo extremely late replication in the cell cycle, even in M-phase by Mitotic DNA Synthesis (MiDAS) as described by Hickson and colleagues . This late replication is the best candidate for the association of CFSs with CNV formation. In contrast, our work, supported by analysis from other labs, showed that R-loop formation was not abundant at these loci and thus did not appear likely to be a main contributor to their instability.
CNV induction by transcription-dependent replication delay. CNV hotspots form when transcription of extremely large genes leads to a prolonged replication delay under stress (top). Poorly described repair mechanisms lead to replication rescue or CNV formation (bottom).
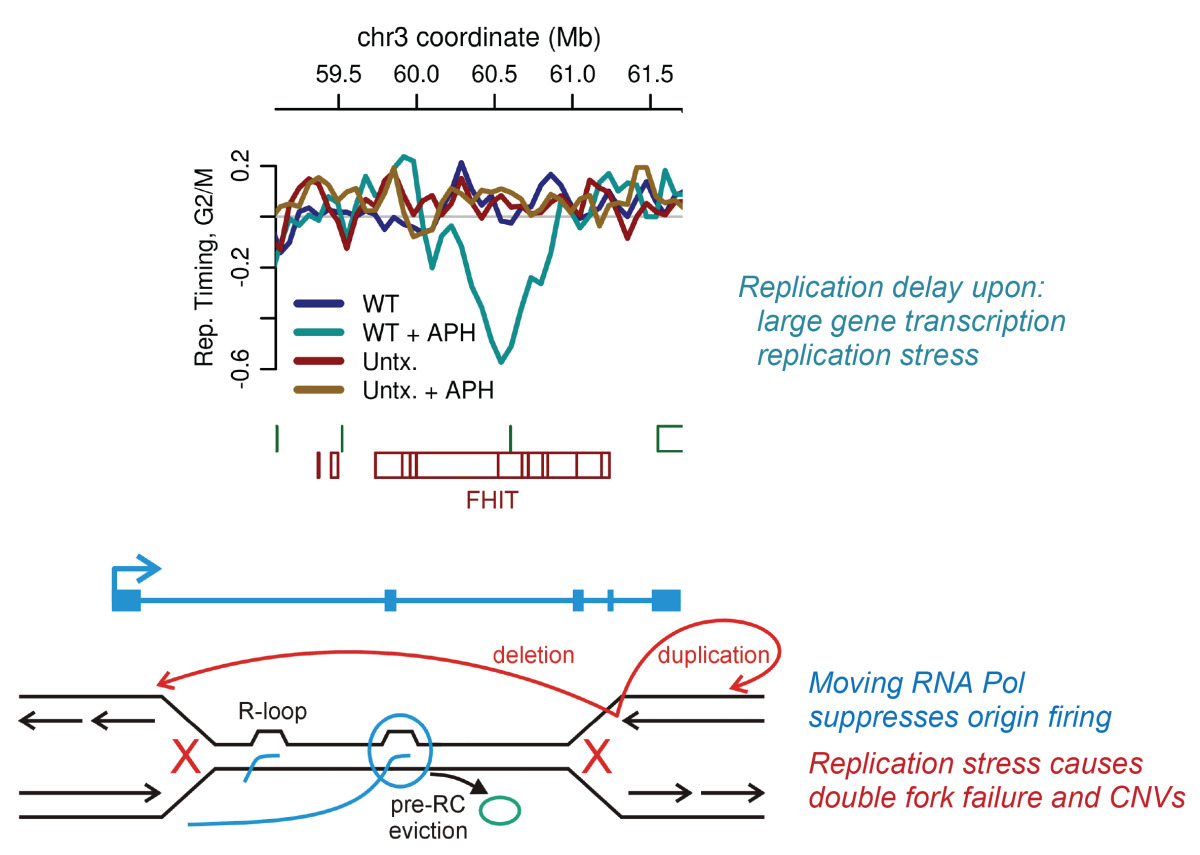
CNV induction by transcription-dependent replication delay. CNV hotspots form when transcription of extremely large genes leads to a prolonged replication delay under stress (top). Poorly described repair mechanisms lead to replication rescue or CNV formation (bottom).
Most recently, we developed a novel genome sequencing approach, svCapture, that allows us to confidently measure the rate of replication-stress-induced SV formation in cells as the junctions form - when there can only ever be one DNA copy of each novel junction . We applied the technique to timed cell cultures to show that induced SVs form primarily after cells enter into mitosis, in a manner dependent on DNA Pol Theta (PolQ)-mediated end joining (TMEJ, also called MMEJ) .
Replication-stress-induced SV induction on entry into mitosis. Remarkably, despite the stress occurring in S-phase, SVs do not form in cells until TMEJ-dependent processing in M.
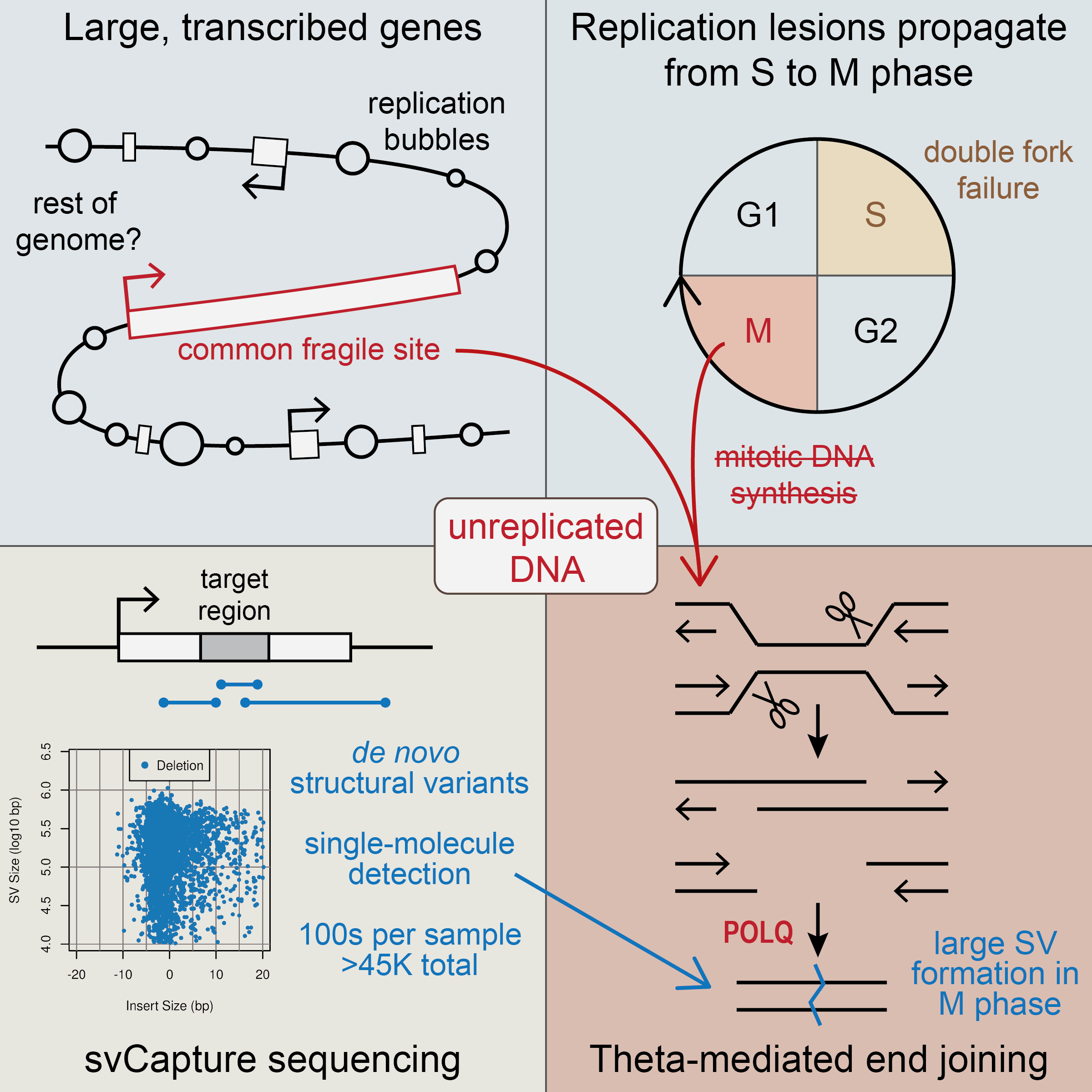
Replication-stress-induced SV induction on entry into mitosis. Remarkably, despite the stress occurring in S-phase, SVs do not form in cells until TMEJ-dependent processing in M.
This line of investigation is important in the context of increasing literature linking chromosomal rearrangements in diseases like cancer to M-phase. SVs at large CFS genes may also play a key role in brain function as it is where those genes are transcribed. Our work highlights the ways in which even subtle alterations in DSB management throughout the cell cycle might lead to increase and/or more deleterious chromosomal outcomes in specific cells, tissues, and people.
Goals and directions
Future goals are to:
- better understand the precise mechanisms by which DSBs are formed at CFS loci that lead to CNV/SV junction formation
- establish the biological significance of mutations formed at CFS genes in vivo
- extend the concepts learned at CFS loci to the rest of the genome through continued advances in associated genomic technologies